R is one of popular scripting language for biologists and often used in the bioinformatics and statistical analysis. The R language has developed several novel packages and libraries, and they are available for academic use without charges. This chapter will guide you to be familiar with basic R syntaxes.
Students are recommended to write R codes by using the RStudio program. There are two modes for the R scripting, the R console (an interactive mode) and the editing mode which can write and save multiple lines of the codes. Codes written in the R editor can be saved in the .R format for later editing.
To begin the R coding, you can start with the set of variables to stores values. Either single and multiple values are able to be assigned to the R variables. A number of special objects are also provided for the storage for specific data types such as character, string, vector, array, list, data frame, etc.
The basic mathematical operation can be executed easily in R using the math operators (+, -, *, /, //, %, ^). The output of the calculation can be kept or assigned to the variable. The variable names must not begin with the number and the names are case sensitive.
Example 1 Yeast has its cell cycle duration of 120 min. If the yeast cells are treated with the fungicide and different mitotic cell stages are counted for the control and treatment.
Control: interphase 180 cells, prophase 9 cells, metaphase 7 cells, anaphase 2 cells, and telophase 2 cells
Treatment: interphase 187 cells, prophase 5 cells, metaphase 6 cells, anaphase 1 cell, and telophase 1 cell
The mitotic index (MI) can be calculated from the number of dividing cells/total number of counted cells as shown by the codes below.
control.total = 180+9+7+2+2
control.mitosis = 9+7+2+2 treatment.total = 187+5+6+1+1
treatment.mitosis = 5+6+1+1 control.MI = control.mitosis/control.total
control.MI treatment.MI = treatment.mitosis/treatment.total
treatment.MI paste(“%MI of control = “, control.MI*100)
paste(“%MI of treatment = “, treatment.MI*100)
Question 1 If the duration of any stage in the cell cycle (min) can be calculated from the number of cells in the stage divided by the total cell number and multiplied with the cell cycle duration of the yeast. Can you show the calculation for all stages in both control and treatment using the R codes?
Question 2 Can you store all values of control and treatment in two variable named control.vector and treatment.vector? What will be the output when execute control.vector == treatment.vector? Please explain the output.
Question 2 Please calculate the output of this equation using R when a = 1, b = 2, and c = 3. The output has to be in two-decimal point format.
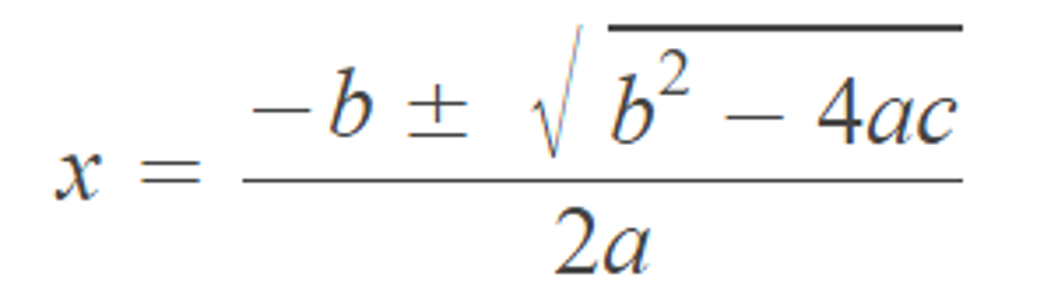
The R language has several functions (paste(), nchar(), length(), strsplit(), substr(), etc) for operating with character and string (a sequence of characters or text). These functions allow to index position in the string, format the string, split and join the string components.
Example 2 Character and string operation
dna.nucleotide = c(‘A’, ‘T’, ‘G’, ‘C’)dna.nucleotide[1]
dna.nucleotide[1:2] rna.nucleotide = dna.nucleotide
rna.nucleotide[2] = ‘U’
rna.nucleotide
rna.nucleotide == dna.nucleotide
paste(dna.nucleotide[1], dna.nucleotide[2], dna.nucleotide[3], sep=””)
DNA = “ATGATGATGTAGTGATGATGAT”length(DNA)
nchar(DNA)
substr(DNA, 1, 5)
DNA.vector = c(“ATGATGATGTAGTGATGATGAT”, “CCCCCCCCCCCGG”)DNA.vector[1] dna.string = paste(DNA.vector[1], DNA.vector[2], sep = “”)
strsplit(dna.string, split = “”) dna.string.split = strsplit(dna.string, split = “”)
dna.string.split
dna.string.split[[1]][1]
Question 3 Can you store this amino acid sequence (MGYINVFAFPFTIYSLLLCRMNSRNYIAQVDVVNFNLT) in an R variable? Please write R codes to split this amino acid sequence at the N amino acid position.
In R, there are several special objects to work with the data. Some of them will be introduced in this chapter. The matrix format is used to store and handle two-dimensional data with rows and columns. Several functions and operations are available. If the data are multi-dimensional, the array object will be more appropriate. The example below shows the preparation of the DNA substitution matrix.
Example 3 Creating a matrix in R
dnasub.vector = c(0.81, 0.10, 0.07, 0.02, 0.07, 0.87, 0.03, 0.03, 0.16, 0.12, 0.71, 0.01, 0.07, 0.26, 0.05, 0.63)
dnasub.matrix = matrix(dnasub.vector, nrow = 4)dnasub.matrix rownames(dnasub.matrix) = c(‘A’, ‘T’, ‘G’, ‘C’)
colnames(dnasub.matrix) = c(‘A’, ‘T’, ‘G’, ‘C’)
dnasub.matrix
dim(dnasub.matrix)
dnasub.matrix[1,1]dnasub.matrix[1:2,1:4] for(i in 1:4){
print(dnasub.matrix[i,1:4])
} for(i in 1:4){
print(dnasub.matrix[1:4, i])
} for(i in 1:4){
for(j in 1:4){
print(dnasub.matrix[i, j])
}
}
Question 4 The following figure is the amino acid substitution matric named BLOSUM62. Please create a matrix to store the data of this BLOSUM matrix.
To the end of the third example, the for statement is the codes for repetitive/loop work (running the same code multiple times). Another repetitive statement is the while loop which differs from the for loop in that it requires the increment or decrement parameter to stop the loop execution. The below example demonstrates the use of the while loop to calculate nucleotide substitution scores when compared two nucleotide sequences with the same length. The given codes also show the use of nested functions (function within function) in R such as print(paste(…)) which helps writing more complex and short codes.
Example 4 The use of while loop
DNA1 = ‘ATGG’DNA2 = ‘AGTC’ DNA.test1 = strsplit(DNA1, split = “”)
DNA.test1 DNA.test2 = strsplit(DNA2, split = “”)
DNA.test2
dnasub.matrix[‘A’,]
DNA.test1[[1]]DNA.test1[[1]][1]
nchar(DNA1)
n = 1
while(n <= nchar(DNA1)){print(paste(“position”, n, ” score = “, dnasub.matrix[DNA.test1[[1]][n], DNA.test2[[1]][n]]))
n = n+1
}
Question 5 Can you write R codes to calculate the total score from the summation of the position score? Please modify the given codes.
For making a conditional statement, you can use the if, if-else or if-else if-else statements to do this. The program will divide which branch of the codes to proceed by checking if the condition is TRUE or FALSE. The TRUE or FALSE is the restricted and reserved words in R. They are Boolean constants and have to be used in all capital letters.
Example 5 The use of conditional statement
amino.full = c(“Ala”, “Ile”, “Leu”, “Met”, “Val”, “Phe”, “Trp”, “Tyr”, “Asn”, “Cyc”, “Gln”,“Ser”, “Thr”, “Asp”, “Glu”, “Arg”, “His”, “Lys”, “Gly”, “Pro”)
amino.short = c(‘A’, ‘I’, ‘L’, ‘M’, ‘V’, ‘F’, ‘W’, ‘Y’, ‘N’, ‘C’, ‘Q’, ‘S’, ‘T’, ‘D’, ‘E’, ‘R’, ‘H’, ‘K’, ‘G’, ‘P’ )
length(amino.full)length(amino.short)
test.protein = strsplit(“ATGCHY”, split = “”)
amino.short[1] == ‘A’test.protein[[1]][1] == amino.short[1]
length(test.protein[[1]])
output.protein = c()
for(i in 1:length(test.protein[[1]])){for(j in 1:20){
if((test.protein[[1]][i] == amino.short[j]) == TRUE){
output.protein[i] = amino.full[j]
}
}
}
output.protein
Question 6 Please modify the given codes and use to calculate the mass of each amino acid. Also if you can add more codes to the program to calculate the total mass of the protein sequence which is calculated from the summation of the mass of all amino acids in the sequence.
