Asst. Prof. Teerasak E-kobon
18 Jan 2021
Several modules in Python facilitates bioinformatics data analysis because of the reusable attributes and functions. Using Python modules reduces programming time and effort to develop new and complicated programs. The built-in or standard modules are available after the language installation (import os, import sys) and use the import statement to access the module or functions within the module. The module can be imported as a different name (import os as basic) or some functions of the module can be imported (from os import getcwwd). Other external or custom functions and modules can be created and installed in the Python environment. A collection of similar modules is named a package. The package contains a ___init__.py file that tells Python to treat the containing directories as the package.
The codes below are the custom function named protcharge() to calculate the protein net charge from the protein sequence input. Once created, you only call the protcharge() function instead of writing the entire scripts again. The argument of the function can be determined (aa_seq), set as the default values, or undetermined numbers with the (*). The line after the def statement is called docstring useful for adding an explanation of the function.
Example 1 Custom function to calculate the protein net charge
def protcharge(aa_seq):
“””Returns the net charge of a protein sequence”””
protseq = aa_seq.upper()
charge = -0.002
aa_charge = {‘C’:-.045, ‘D’:-.999, ‘E’:-.998, ‘H’:.091, ‘K’:1, ‘R’:1, ‘Y’:-.001}
for aa in protseq:charge += aa_charge.get(aa,0)
return charge
Question 1 Can you run and test this function? Please show the output.
There are several ways to install external modules in Python. To begin this section, the PyCharm editor is recommended to use in this chapter. This code editor can be used for both Python and R programming. The bottom tab of this editor has OS terminal, Python and R consoles accessible for the interactive mode coding (Figure 1).
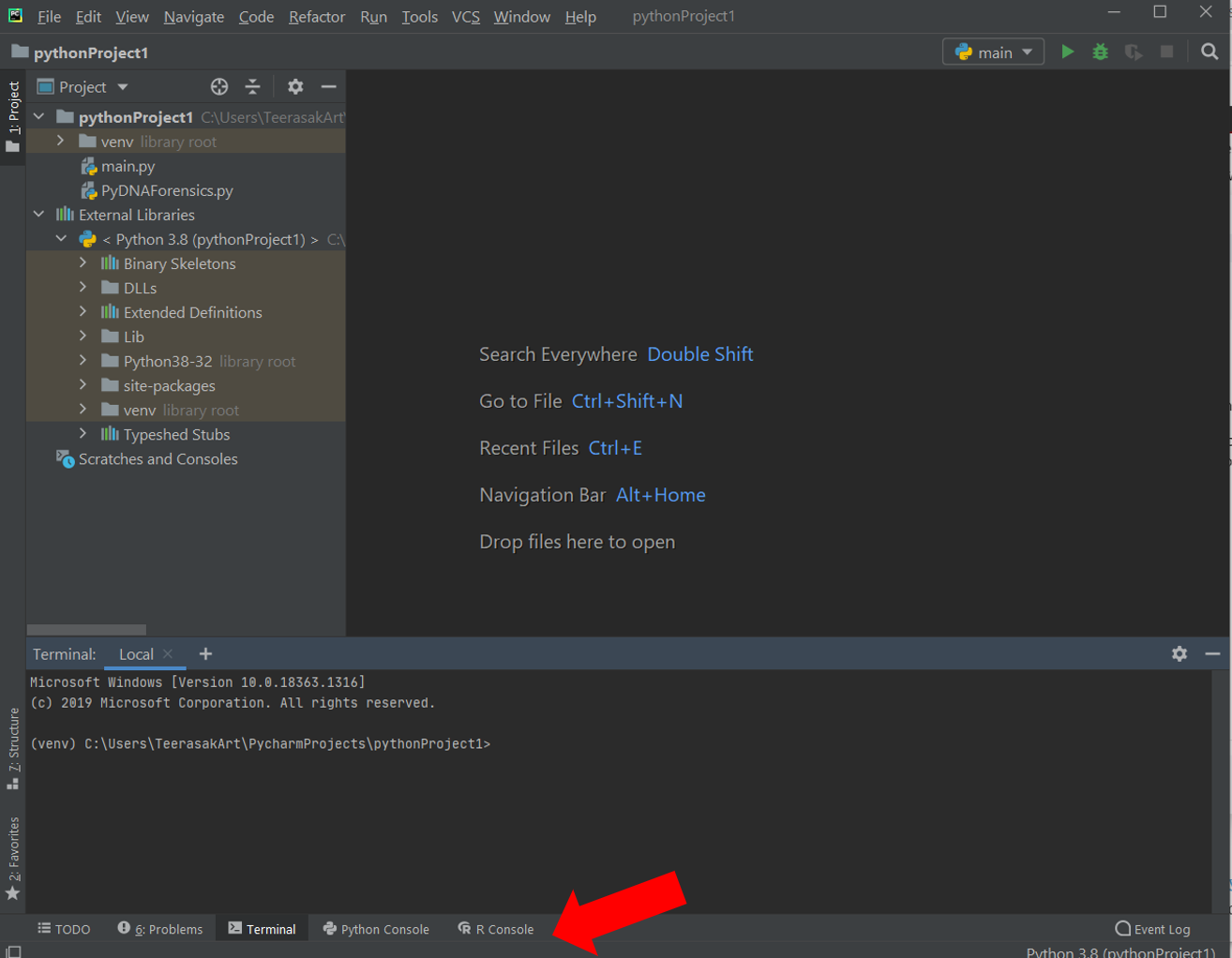
Figure 1 PyCharm editor
The first method to install modules in Python is using the pip install command followed by the module name (Figure 2). The use of pip command in the PyCharm terminal is happened through the virtualenv program to create the isolated Python environment for each program. The second method is to use the conda install command in the Anaconda virtual environment. This will not be demonstrated in the chapter as it was taught previously. The third method is to manually build and install the modules.
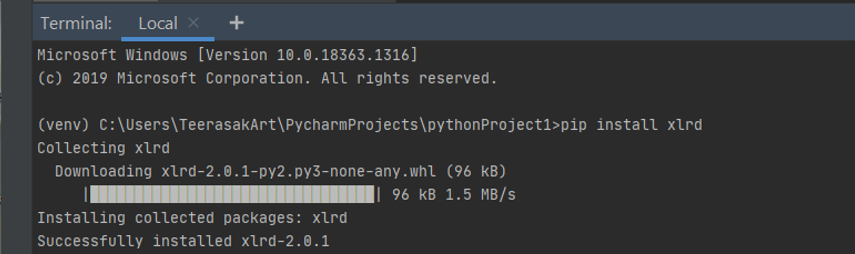
Figure 2 Using pip command to install a new Python module
Information on available public and shared Python modules can be searched by reading the bioinformatics research articles such as the Oxford Bioinformatics Journal or the code repository (GitHub). The second example below is obtained from the publication of Buchmann and Holmes (2019) who built the Python Package to interact with the NCBI Entrez system. This module can be installed by using pip install entrezpy command. The connection to the NCBI database might need the NCBI API-key following the provided guideline.
Example 2 Use of the Entrezpy module
import entrezpy.esearch.esearcher e = entrezpy.esearch.esearcher.Esearcher(‘esearcher’, ’email’) a = e.inquire({‘db’:’nucleotide’,’term’:’viruses[orgn]’, ‘retmax’: 10, ‘rettype’: ‘uilist’}) print(a.get_result().uids)Question 2 Can you install and try to use this EntrezPy module with the given examples? More tutorials on the module are available. Can you modify the given codes to search the sequence IDs from other organisms in the NCBI database?
The last method is to create your own module by creating the .py file and saving the file in the directory where the Python interpreter can find it.
Question 3 Please create a new Python module for calculation of the protein isoelectric pH and demonstrate the codes to import and run.
Sometimes, the codes are developed by other programmers and shared in their GitHub repository. The users can download and re-use their developed codes in the current Python program. The simplest way to do is to open the borrowed module file and run under your current environment. The third example is the Entrez module developed by Jordi Burguet-Castell to query and retrieve the required data from the NCBI database. Please download this module and try the following codes. Hint: the example below only use the entrez.py file.
Example 3 The use of the Entrez module available from the GitHub repository
import entrez # Fetch information for SNP with id 3000 for line in entrez.equery(tool=’fetch’, db=’snp’, id=’3000′): print(line) # Get a summary of nucleotides related to accession numbers NC_010611.1 and EU477409.1 for line in entrez.on_search(db=’nucleotide’, term=’NC_010611.1[accn] OR EU477409.1[accn]’, tool=’summary’): print(line) # Download all chimpanzee mRNA sequences in FASTA format # This codes will take times to run. with open(‘C:\Users\TeerasakArt\Downloads\chimp.fna’, ‘w’) as fout: for line in entrez.on_search(db=’nucleotide’, term=’chimpanzee[orgn] AND biomol mrna[prop]’, tool=’fetch’, rettype=’fasta’): fout.write(line + ‘n’)Question 4 Can you modify the codes to use the Entrez module to download 10 nucleotide sequences of the S gene of the COVID-19 virus?
The last module to introduce in this chapter is the BioPython which has several useful modules and packages for bioinformatics analysis. The BioPython Wiki also provides several useful guidelines to use this package. This module can be installed by the pip install biopython command and check successful import by import Bio statement on the Python terminal. The BioPython creates specific objects for biological sequences (Seq object) and sequence file formats (SeqIO object). You can import the Seq module to call several useful functions such as complement(), reverse.complement(), find(), count(), transcribe(), back_transcribe(), and translate(). The Seq.IO object also allows you to parse several sequence file formats (FASTA, GenBank) with the parse() function for the latter bioinformatics analysis without worrying on the file formatting. The code examples are available in the BioPython document.
Question 5 Can you use the Seq and SeqIO modules to read a FASTA file that contains a single nucleotide sequence of the S gene of the COVID-19 virus and convert it to the complementary strand and amino acid sequence?
****************
